Article of the week: Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: a 23‐gene panel with utility for non‐invasive diagnosis and risk stratification
Every week, the Editor-in-Chief selects an Article of the Week from the current issue of BJUI. The abstract is reproduced below and you can click on the button to read the full article, which is freely available to all readers for at least 30 days from the time of this post.
In addition to the article itself, there is an editorial written by a prominent member of the urological community and a video prepared by the authors. These are intended to provoke comment and discussion and we invite you to use the comment tools at the bottom of each post to join the conversation.
If you only have time to read one article this week, it should be this one.
Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: a 23‐gene panel with utility for non‐invasive diagnosis and risk stratification
Douglas G. Ward*, Naheema S. Gordon*, Rebecca H. Boucher*, Sarah J. Pirrie*, Laura Baxter†, Sascha Ott†, Lee Silcock‡, Celina M. Whalley*, Joanne D. Stockton*, Andrew D. Beggs*, Mike Griffiths§, Ben Abbotts*, Hanieh Ijakipour*, Fathimath N.Latheef*, Robert A. Robinson*, Andrew J. White*, Nicholas D. James*, Maurice P.Zeegers¶, K. K. Cheng** and Richard T. Bryan*
*Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham, †Department of Computer Science, University of Warwick, Coventry, ‡Nonacus Limited, Birmingham Research Park, §West Midlands Regional Genetics Laboratory, Birmingham Women’s and Children’s NHS Foundation Trust, Birmingham, UK, ¶NUTRIM School for Nutrition and Translational Research in Metabolism and CAPHRI Care and Public Health Research Institute, Maastricht University, Maastricht, The Netherlands and **Institute of Applied Health Research, University of Birmingham, Birmingham, UK
Abstract
Objectives
To develop a focused panel of somatic mutations (SMs) present in the majority of urothelial bladder cancers (UBCs), to investigate the diagnostic and prognostic utility of this panel, and to compare the identification of SMs in urinary cell‐pellet (cp) DNA and cell‐free (cf) DNA as part of the development of a non‐invasive clinical assay.
Patients and Methods
A panel of SMs was validated by targeted deep‐sequencing of tumour DNA from 956 patients with UBC. In addition, amplicon and capture‐based targeted sequencing measured mutant allele frequencies (MAFs) of SMs in 314 urine cpDNAs and 153 urine cfDNAs. The association of SMs with grade, stage and clinical outcomes was investigated by univariate and multivariate Cox models. Concordance between SMs detected in tumour tissue and cpDNA and cfDNA was assessed.
Results
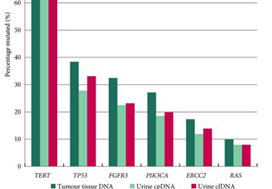
The panel comprised SMs in 23 genes: TERT (promoter), FGFR3, PIK3CA, TP53, ERCC2, RHOB, ERBB2, HRAS, RXRA, ELF3, CDKN1A, KRAS, KDM6A, AKT1, FBXW7, ERBB3, SF3B1, CTNNB1, BRAF, C3orf70, CREBBP, CDKN2A and NRAS; 93.5–98.3% of UBCs of all grades and stages harboured ≥1 SM (mean: 2.5 SMs/tumour). RAS mutations were associated with better overall survival (P = 0.04). Mutations in RXRA, RHOB and TERT (promoter) were associated with shorter time to recurrence (P < 0.05). MAFs in urinary cfDNA and cpDNA were highly correlated; using a capture‐based approach, >94% of tumour SMs were detected in both cpDNA and cfDNA.
Conclusions
SMs are reliably detected in urinary cpDNA and cfDNA. The technical capability to identify very low MAFs is essential to reliably detect UBC, regardless of the use of cpDNA or cfDNA. This 23‐gene panel shows promise for the non‐invasive diagnosis and risk stratification of UBC.