Editorial: Shift from protocol-based to personalized medicine in active surveillance: beginning of a new era
The use of active surveillance (AS) is rapidly expanding worldwide, with rates as high as 74% among patients with low-risk prostate cancer in the nationwide registry of Sweden [1]. Despite increasing uptake of this strategy by patients, there is no consensus among the medical community as to the ideal criteria for selection and monitoring [2]. For example, the Johns Hopkins AS programme restricts enrolment to men with low-risk disease and performs annual biopsies for monitoring. Other protocols also include men with intermediate-risk disease and perform prostate biopsy at less frequent intervals.
Is it really optimal to use the same follow-up protocol for all patients? Many factors influence the risk of reclassification, including patient characteristics (e.g. race, body mass index) and disease features (e.g. PSA density, Gleason score and extent of disease on biopsy) [3]. Moreover, previous studies have shown that the risk of reclassification during AS is a conditional probability, where the risk decreases with each additional negative biopsy [4]. Given that individual patients have vastly different risks of reclassification, and that the risk changes over time, AS represents an ideal context for personalized medicine.
There has already been a significant paradigm shift in prostate cancer screening from a one-size-fits-all to a multivariable, risk-adapted approach [5]. Why would we use the same screening intervals and biopsy cutoff for patients with vastly different risk profiles? Multiple guidelines already recommend using PSA levels to guide screening protocols, and there are several validated multivariable tools to provide more personalized estimates of prostate cancer risk. Both the Prostate Cancer Prevention Trial (PCPT) and the European Randomised Study of Screening for Prostate Cancer (ERSPC) risk calculators have been extensively studied and are readily available online for use in clinical practice [6].
To date, the concept of risk-adapted AS has received relatively little attention, and few nomograms have been created specifically for the AS population. Using data from the Canary Prostate Active Surveillance Study (PASS), Ankerst et al. [7] designed a nomogram to predict biopsy reclassification using age at biopsy, months since the last biopsy, last PSA level, percentage of cores positive for cancer on the last biopsy, and number of previous negative biopsies. This tool had an area under the curve (AUC) of 0.724 on internal validation, and is available online at https://prostate-cancer-risk-calculator.org to facilitate additional validation and clinical use.
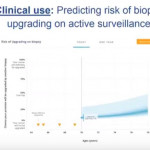
In the current issue of BJUI, Mamawala et al. [8] report on the development of another new AS nomogram using data from the Johns Hopkins programme. Specifically, the tool predicts the risk of biopsy reclassification using six variables: age; PSA density; year of diagnosis; laterality; risk strata; and total number of biopsies. The nomogram was well calibrated and had an AUC of 0.757 on internal validation. Notably, the same authors have also recently developed a different tool to predict pathological Gleason score for men on AS using a Bayesian joint model [9]. Following external validation, these tools may help provide more customized decision support for the AS population by integrating longitudinal data.
It is noteworthy that none of these nomograms incorporate new markers or imaging, and it is likely that such data could further refine their estimates. For example, longitudinal measurements of the Prostate Health Index were previously shown to predict biopsy reclassification during AS [10], and the use of multiparametric MRI continues to expand. As more data on these tests become available, the AS risk calculators should be updated, as has been done with the PCPT and ERSPC risk calculators used in the screening context. In the future, continued research on genetics may allow further tailoring of AS. In the meantime, these risk calculators are an important first step (‘version 1.0’) toward a more personalized approach to AS.